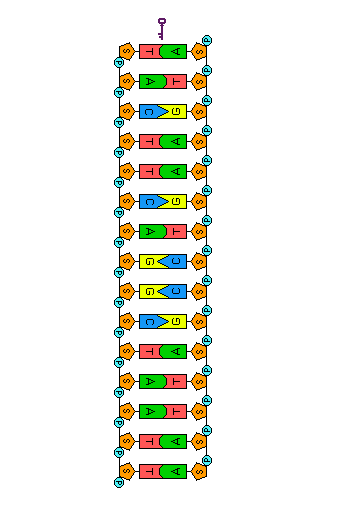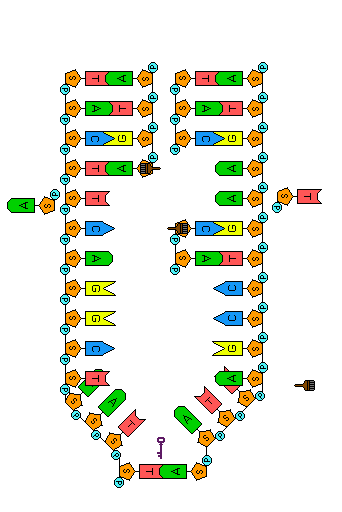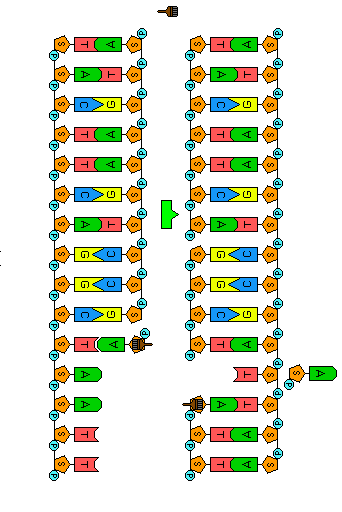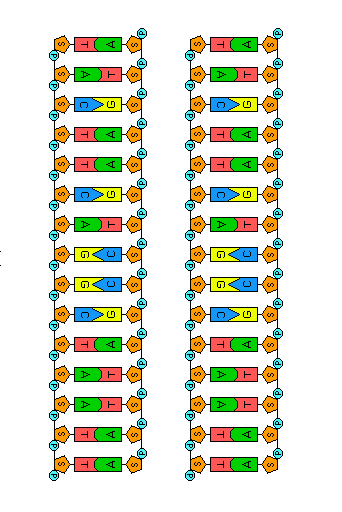
Before a cell divides, its DNA must be replicated (duplicated). Because the two strands of a DNA molecule have complementary base pairs, the nucleotide sequence of each strand automatically supplies the information needed to produce its new partner. The two strands of a DNA molecule are separated and each is used as a pattern or template to produce a complementary strand. Each template and its new complement together then form a new DNA double helix, identical to the original. Because the two new DNA molecules each contain a new strand of nucleotides along with one strand from the parent molecule, we say that the process of DNA replication is semi-conservative.
Before replication can occur, the length of the DNA double helix about to be copied must be unwound. Once the DNA strands have been unwound, they must be unzipped to expose the bases so that new nucleotide partners can bond to them. These tasks are accomplished by the enzyme helicase—one of many proteins that work as a micro-tools inside the cell. In the animation to the left, helicase is represented by the small key to symbolize unlocking the molecule.