Sanger Sequencing Video I
Yourgenome (28 Sep 2016) DNA Sequencing - 3D [Video file] retrieved from https://https://youtu.be/ONGdehkB8jU
Yourgenome (28 Sep 2016) DNA Sequencing - 3D [Video file] retrieved from https://youtu.be/ONGdehkB8jU
DNA is a miraculous macromolecule that carries the instructions for the processes of life from generation to generation of living organism. The goal of DNA research is to decode and understand those instructions. We know that DNA codes for the order of amino acids in all the proteins of a living organism. By the 1960’s it was well established that a sequence of three nucleotide bases in a length of DNA (a codon) codes for one amino acid. During that time, the code for each nucleotide triplet pair and its corresponding amino acid was worked out as well. Soon researchers were reporting DNA sequences that coded for some of the smaller proteins and some RNA molecules. However, it wasn’t until 1995 that the first genome of a living organism was completely sequenced. That was the 1.8 million base pairs of the bacterium Haemophilus influenza.
Well before the sequencing of H. influenza, in 1990, scientists around the world had come together to launch the Human Genome Project. The Human Genome Project was an ambitious project designed to produce a complete and accurate sequence of the 3 billion base pairs that make up the DNA of the Human Genome. The second goal of the project was to identify all the genes within the sequence of human DNA. The project was completed ahead of schedule in 2003.
The challenge of applying meaning to these strings of billions of A’s, T’s, G’s and C’s is a project that will stretch far into the future. Scientists are only beginning to associate various strings with the traits they control and to understand the “mistakes” that are responsible for certain illnesses and disorders.
Over the years, different procedures for determining the sequence of bases in DNA molecules have been developed. Early methods were exacting and time consuming and suffered from a lack of adequate computing power to interpret data. One of the methods most commonly used for the Human Genome and other molecular genetics projects is the Sanger Sequencing Method.
Strattti (11 Nov 2015) The Sanger Method of DNA Sequencing [Video file] retrieved from https://youtu.be/FvHRio1yyhQ
Strattti (11 Nov 2015 The Sanger Method of DNA Sequencing [Video file] retrieved from Strattti (11 Nov 2015 The Sanger Method of DNA Sequencing [Video file] retrieved from https://youtu.be/FvHRio1yyhQ
If given the few basic resources it needs, DNA can replicate outside of the living cell (We say replication that occurs outside the cell occur in vitro as opposed to replication that is carried out inside a living cell or in vivo.) After DNA from a sample is isolated, cut into small segments, and amplified (multiplied) by PCR, it is placed in a buffer medium with a mixture of all four free nucleotides, a DNA polymerase, an enzyme responsible for adding new nucleotides to the original DNA strand, and a primer designed to bind to the 5‘ end of the target fragment to be analyzed. The DNA mixture is then divided into four separate containers.
The “special ingredients” of the Sanger sequencing method are modified versions of the four nucleotide bases of DNA called dideoxynucleotides (ddNTPs). Each of the reaction mixtures will receive a small amount of one of the four altered bases.
If given the few basic resources it needs, DNA can replicate outside of the living cell (We say replication that occurs outside the cell occur in vitro as opposed to replication that is carried out inside a living cell or in vivo.) After DNA from a sample is isolated, cut into small segments, and amplified (multiplied) by PCR, it is placed in a buffer medium with a mixture of all four free nucleotides, a DNA polymerase , an enzyme responsible for adding new nucleotides to the original DNA strand, and a primer designed to bind to the 5‘ end of the target fragment to be analyzed. The DNA mixture is then divided into four separate containers.
The “special ingrediants” of the Sanger sequencing method are modified versions of the four nucleotide bases of DNA called dideoxynucleotides (ddNTPs). Each of the reaction mixtures will receive small amount of one of the four altered bases.
Strattti (11 Nov 2015) The Sanger Method of DNA Sequencing [Video file] retrieved from https://youtu.be/FvHRio1yyhQ
Strattti (11 Nov 2015 The Sanger Method of DNA Sequencing [Video file] retrieved from Strattti (11 Nov 2015 The Sanger Method of DNA Sequencing [Video file] retrieved from https://youtu.be/FvHRio1yyhQ
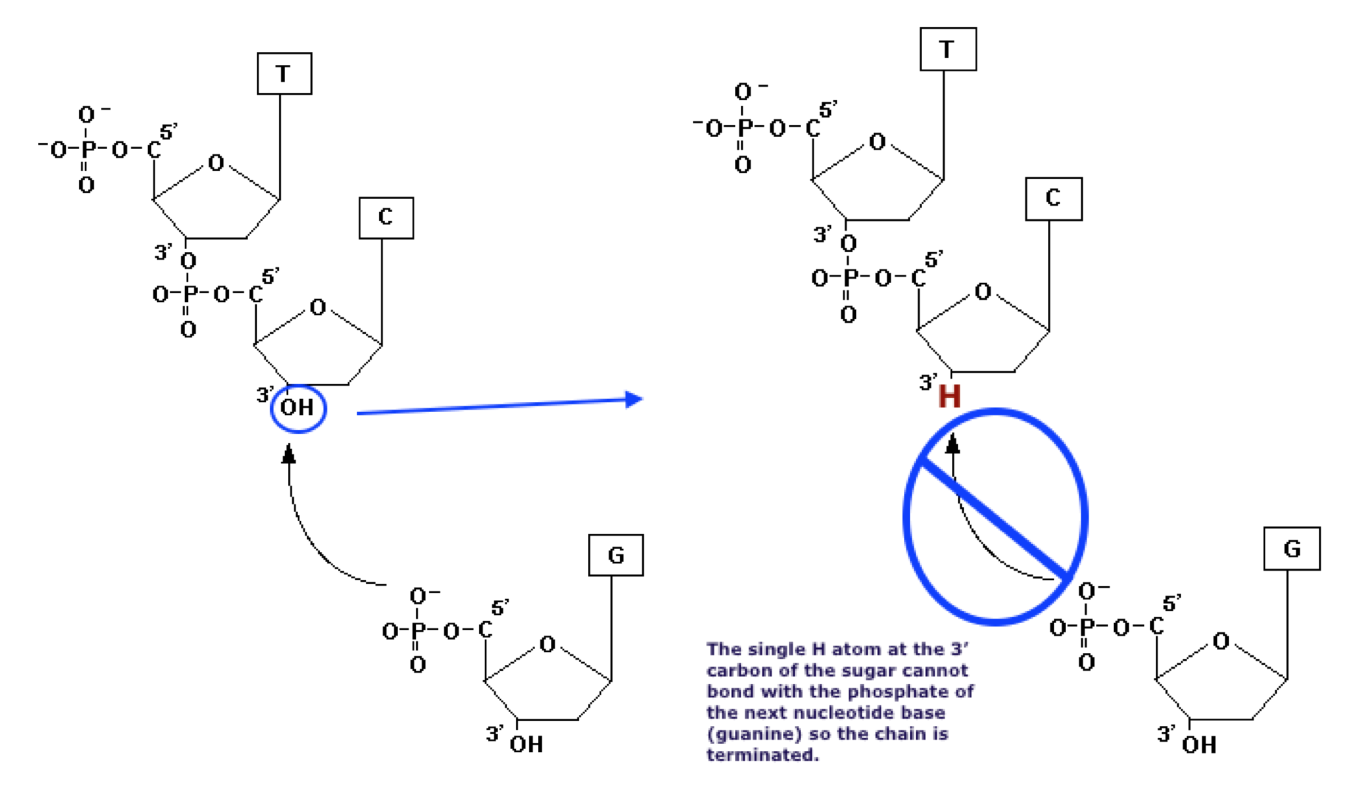

The altered nucleotides will be incorporated randomly into the growing DNA chains. Fragments of DNA built of normal dNTP’s will be extended until a random ddNTP finds a place in the growing chain. The ddNTP’s phosphate group will bind normally to the 5’ carbon of the previous nucleotide. However, the chain cannot be lengthened further because the ddNTP cannot bind with another nucleotide on its altered 3’ end.
After 20 - 30 cycles of the PCR heating and cooling, the resulting mixture will contain a series of fragments of different lengths. The length of each fragment will depend on how many bases had been added to the chain before one of the ddNTPs sneaked in and blocked further growth. Each fragment in a container will be terminated by the same ddNTP. in the tube containing ddATP, all the DNA fragments will end with A. In the mixture with ddGTP, the fragments will end with G and so on.
The mixtures of DNA fragments are nearly transparent and do not show up well on the gel. In the early years of Sanger Sequencing, the DNA bases in the primers were tagged with radioactive phosphorous. After they had been separated by electrophoresis, it was necessary to coat the gel with a photosensitive emulsion (like that on photographic film).
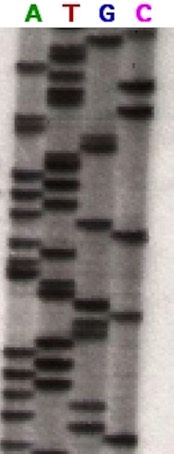
As the emulsion was exposed to the decay emissions of the radioactive phosphorous it produced an image of the distribution of the DNA fragments. The emulsion was developed and the placement of the segments could be viewed as in the figure to the right.
Figure 2 is an autoradiogram of a gel electrophoresis run of four mixtures of the Sanger method. The mixture containing ddATP is on the left. Every segment spread over that lane will have an A in its terminal position. The segments in the second lane were incubated with ddTTP and each segment has a T in the terminal position. Similarly, lanes three and four contain segments ending in G and C respectively. Because smaller fragments migrate through a gel more quickly, the sequence of nucleotide bases in the original DNA segment is read from the bottom up. The bottom of the gel in this image has been cropped. However, assuming all the bars were conserved in the crop, the first segment is in lane 2 telling us that the first base in the target DNA segment was T. The next bar is in the first lane making the second base A. Reading up the gel, the sequence of the target segment is TACGAGATATATGGCGTTAATACGATATATTGGAACTTCTATTGC
After 20 - 30 cycles of the PCR heating and cooling, the resulting mixture will contain a series of fragments of different lengths depending on how many bases had been added to the chain before one of the ddNTPs sneaked in and blocked further growth. Each fragment in a container will be terminated by the same ddNTP. In the tube containing ddATP, all the DNA fragments will end with A. In the mixture with ddGTP, the fragments will end with G and so on.
The mixtures of DNA fragments are nearly transparent and do not show up well on the gel. In the early years of Sanger Sequencing, the DNA bases in the primers were tagged with radioactive phosphorous. After they had been separated by electrophoresis, it was necessary to coat the gel with a photosensitive emulsion (like that on photographic film).
As the emulsion was exposed to the decay emissions of the radioactive phosphorous it produced an image of the distribution of the DNA fragments. The emulsion was developed and the placement of the segments could be viewed as in the figure to the right.
Figure 2 is an autoradiogram of a gel electrophoresis run of four mixtures of the Sanger method. The mixture containing ddATP is on the left. Every segment spread over that lane will have an A in its terminal position. The segments in the second lane were incubated with ddTTP and each segment has a T in the terminal position. Similarly, lanes three and four contain segments ending in G and C respectively. Because smaller fragments migrate through a gel more quickly, the sequence of nucleotide bases in the original DNA segment is read from the bottom up. The bottom of the gel in this image has been cropped. However, assuming all the bars were conserved in the crop, the first segment is in lane 2 telling us that the first base in the target DNA segment was T. The next bar is in the first lane making the second base A. Reading up the gel, the sequence of the target segment is TACGAGATATATGGCGTTAATACGATATATTGGAACTTCTATTGC

By John Schmidt (en:Image:Sequencing.jpg) [GFDL (http://www.gnu.org/copyleft/fdl.html) or CC-BY-SA-3.0 (http://creativecommons.org/licenses/by-sa/3.0/)], via Wikimedia Commons
Scientists worked to refine the sequencing process: to make it faster, cheaper, safer and more efficient. One major improvement was to label each ddNTP’s with a different color fluorescent dye. (The base colors are traditionally labeled so that adenine is green (A), thymine red (T), cytosine is blue (C) and guanine is yellow (though usually shown in images as black—G—since yellow does not show up well in print). This move to fluorescence labels greatly enhanced the safety and convenience of sequencing by eliminating the precautions, risks and expense associated with working with radioactive materials. Additionally this differential labeling allowed the entire sequencing process to be carried out in one mixture containing all four ddNTP’s.
Another advance in Sanger sequencing was the introduction of glass capillary tubes to hold the gel sieving matrix. This allowed more samples to be run at once in a limited area. A laser beam is passed through the tube near the end. Each of the four ddNTP’s fluoresces a different color when illuminated by a laser beam.
An automatic scanner records each wavelength of light and a computer generates an electropherogram with colored peaks that represent the wavelength of the terminal ddNTP in the sequence as it passes through the beam. The 5' terminal base (the ddNTP) of the shortest fragment (that moves the fastest) is the first base in the electropherogram.
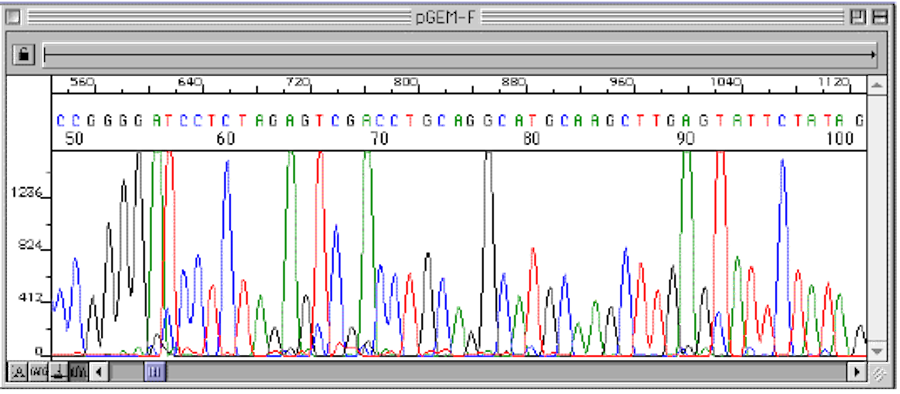
Figure 3 An electropherogram generated by automated Sanger Sequencing showing the nucleotide sequence of a target DNA segment.
The resolution of this process is so powerful that it can show the difference of one nucleotide between two different fragments of DNA.
Figure 3 is a plot of the colors detected in one DNA sample, scanned from the smallest fragments to largest. The computer interprets the colors by printing the nucleotide sequence across the top of the plot.
Figure 2 above:
By John Schmidt (en:Image:Sequencing.jpg) [GFDL (http://www.gnu.org/copyleft/fdl.html) or CC-BY-SA-3.0 (http://creativecommons.org/licenses/by-sa/3.0/)], via Wikimedia Commons