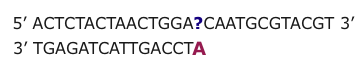Early methods of characterizing DNA were expensive, time consuming and limited in scope. The Sanger Sequencing method was used for most of the Human Genome Project. Over the years, Sanger Sequencing has undergone repeated updates until it is now a highly-automated process that can handle as many as 96 samples of test DNA at a time. Sanger Sequencing is still the gold standard for DNA sequencing. It does not readily lend itself to efficient SNP typing.
In the last ten years or so dramatic advances in technology have made it possible to analyze thousands of different DNA segments simultaneously. There are many variations of these new technologies and many more in development. As a group, they are frequently referred to as Next Generation or Next Gen. Next Generation technologies are especially well suited for SNP characterization.
Next Generation technologies are based on the natural ability of DNA to hybridize. A single strand of DNA nucleotides (ssDNA) can hybridize (match up and form a double strand held together by hydrogen bonds) with any other ssDNA that has a complementary nucleotide sequence. This is true even if the two strands are NOT from the same source.
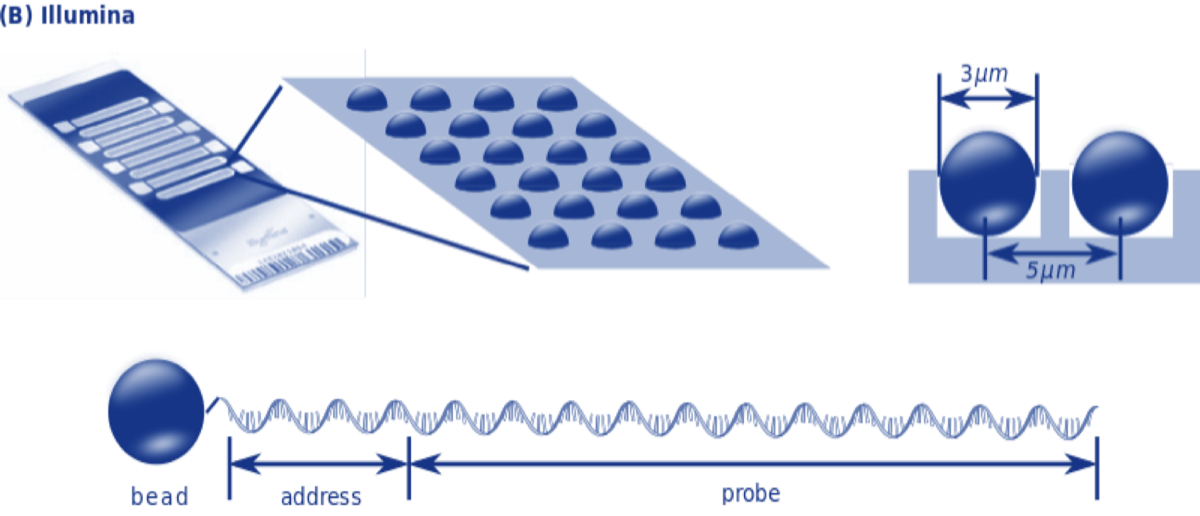
Next Generation technologies typically bind millions of short oligonucleotide fragments (short chains of nucleotides) of known sequences to microscopic dots or minuscule silicon beads on the surface of a small glass or silicon slide. These fragments, called oligos, serve as probes than can hybridize to fragments of unknown DNA.
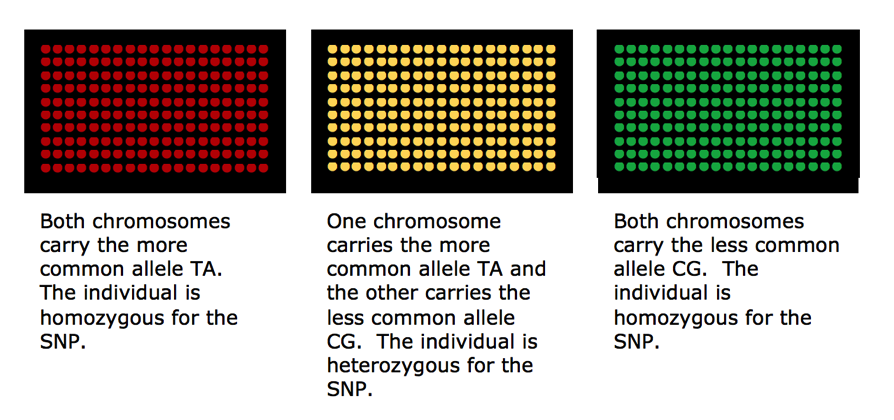
It is then possible to infer the nucleotides of the unknown DNA strand using base pairing rules and knowledge of the nucleotide sequence of the probe.
There are a variety of ways microarray chips can be designed, produced and used depending on research goals and objectives. The three major US companies that provide direct to the consumer DNA SNP testing for genealogy are AncestryDNA, Family Tree DNA and 23andme. All three use some version of the Illumina OmniExpress Chip.